Genetically defined ancestry reveals unique prostate cancer associated polymorphisms in men of African ancestry: analysis of the ELLIPSE Prostate Cancer Cohort
Meghana S. Pagadala, BS1, Joshua A. Linscott, MD, PhD2, Hannah Carter, PhD1, Karim A. Kader, MD, PhD3, Stephen T. Ryan, MD2.
1University of California San Diego School of Medicine, San Diego, CA, 2Maine Medical Center, Portland, ME, 3University of California San Diego Health, San Diego, CA.
BACKGROUND: Prostate cancer (PrCa) is the second leading cause of cancer death in U.S. men and while PSA screening can aid in detection, better tools are needed to stratify risk. Prior Genome-wide association studies (GWAS) using self-reported ancestry have identified PrCa associated Single Nucleotide Polymorphisms (SNPs), but the majority of these findings are specific to patients of European ancestry. We hypothesized that genetically defined ancestral groups would be a more accurate assessment of PrCa associated SNPs.
METHODS: Genetic, phenotype, and self-identified ethnicity (European, African American, Latino, Asian) data was obtained from the ELLIPSE Prostate Cancer Meta-Analysis and Genotyping study (a case-control study of over 99,000 men). SNPs were imputed with the Michigan Imputation Server using the 1000 Genomes Project as the reference population. Genetic ancestry analysis was conducted using a merged identity-by-state matrix (ELLIPSE and HAPMAP study) and Principal Component Analysis (PCA). A k-means clustering model trained on HAPMAP data was used to determine that the ELLIPSE cohort best fit with 5 ancestral groups from the HAPMAP data: European n=82551, African n=6355, Mexican n=1925, Indian n=522 and Asian n=291. Self-identified and genetically defined ancestral groups were compared with PCA. Genome-wide analysis were performed and SNPs passing the p-threshold of 5x10-8 were analyzed.
RESULTS: Genetically defined ancestral groups better controlled for effects of population stratification than self-identified ancestral groups (p<3.47x10-9, Figure 1). European and African ancestral groups were well-powered for analysis with numerous SNPs passing the p-threshold. Manhattan plot of the African ancestral group showed significant associations on chromosome 8, with multiple SNPs aligning with described loci in European populations. Unique SNPs were noted on chromosomes 6 and 11 (Figure 2). The Mexican, Indian and Asian ancestral groups were not sufficiently powered for analyses.
CONCLUSIONS: Patients in the ELLIPSE cohort have significantly better grouping when stratified by genetic ancestry compared to self-identified ancestry. Specifically, the cohort can be sorted into 5 ancestral groups that align with groups described by the International HAPMAP Project. Furthermore, GWAS using genetically identified ancestry uncovered SNPs specific to minority ethnic groups. Future characterization of these PrCa risk SNPs could aid individualized screening recommendations within the African American population. 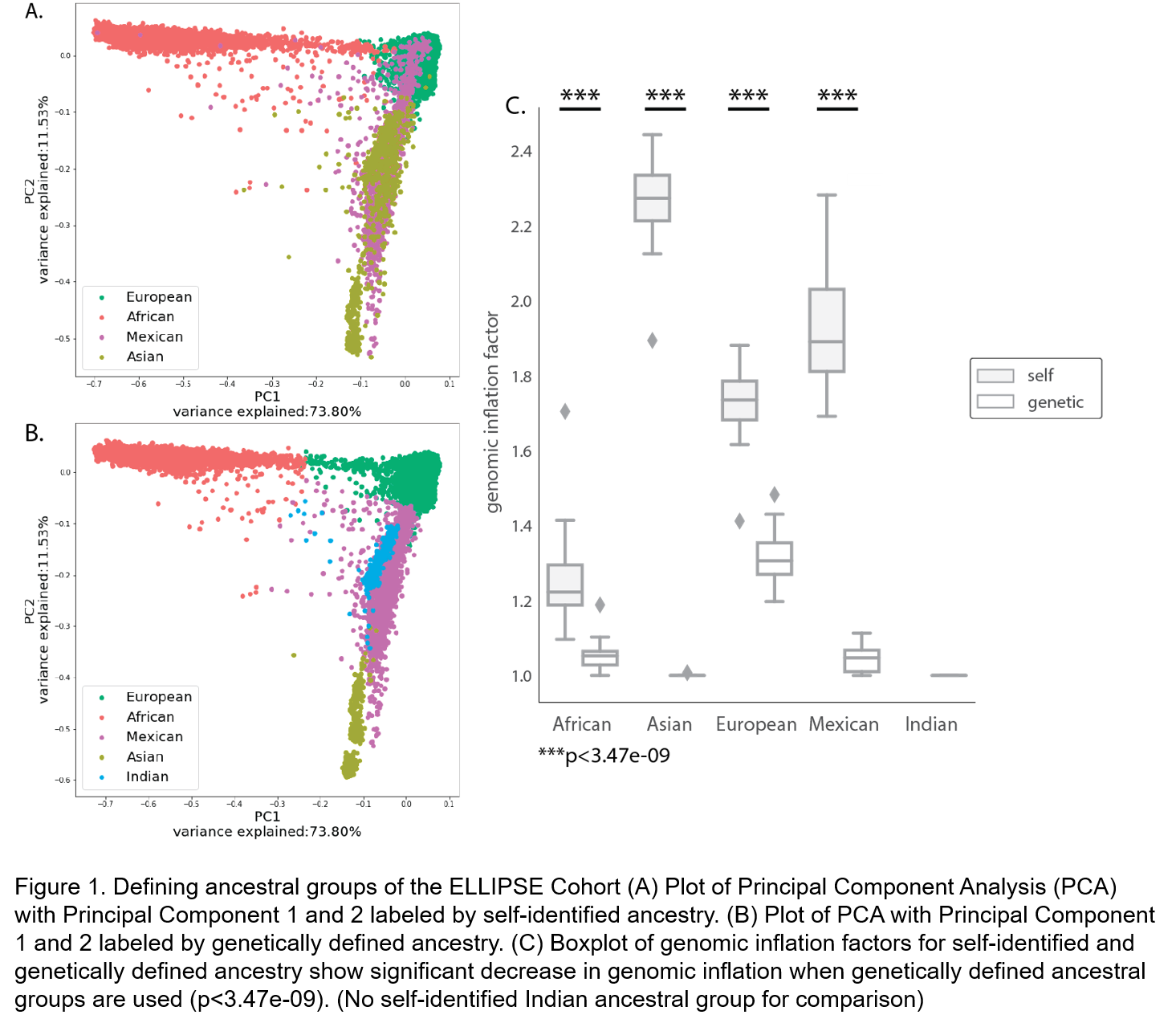

Back to 2020 Abstracts
