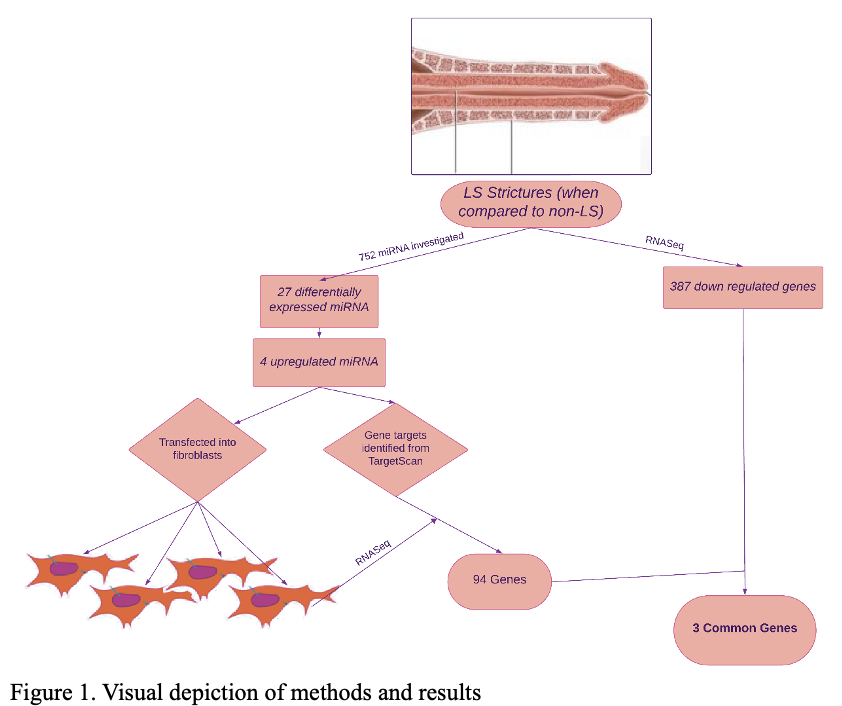
BACKGROUND: The pathophysiology of lichen sclerosus urethral strictures (LS-US) is poorly understood. Previous work has identified 27 differentially expressed miRNA between LS and non-LS urethral stricture (US). The goal of this study was to identify genes that may be downregulated by miRNA associated with LS-US. METHODS: In a previously published study, 27 differentially expressed miRNA in LS-US were identified. The 4 highest upregulated miRNA in LS samples were miR-155-5p, -150-5p, -146-5p, and -142-3p. Through transfection, we forced expression of these miRNA in two foreskin fibroblast cell lines (HDFn and HDFa) and subsequently performed RNA-seq analysis to identify downregulated genes (p<0.1). The identified genes were compared to theoretical gene targets of the 4 miRNA identified using TargetScan.In a concurrent study, RNA-seq analysis was performed on LS and non-LS US tissue (n total =82) to identify genes that were dysregulated in LS (FDR <0.001). These genes were crossed referenced with the gene targets of the 4 different miRNA to identify consensus genes (Figure 1).We also transfected fibroblast cultures with the 3’UTR of candidate genes and performed luciferase assays to assess the impact on the target genes. RESULTS: Comparing the potential miRNA targets from TargetScan and the fibroblast miRNA transfection data, 94 common genes were identified. In the US sequencing data, 387 genes were downregulated in LS tissue. The 94 miRNA target genes were compared to the 387 downregulated genes and 3 genes were in common. For two of these gene products there was a significant 2-fold downregulation in both the LS-US and in the transfected fibroblasts. However, there have yet to be any identified links between these genes and LS or conditions with similar pathophysiology. TGFB2 was downregulated 2.2-fold in LS-US (p= 2.7E-08) and 2.5-fold in the transfected fibroblasts (p= 0.086). The luciferase assay confirmed binding of miR-142-3p with the 3’UTR of TGFB2. Transfected fibroblasts with a wild-type 3’UTR exhibited an 18% decrease in TGFB2 expression (p=0.009) while the mutated version did not have any significant change in TGFB2 expression. This gene has been identified in previous studies investigating juvenile LS, vulvar LS, and SCC. CONCLUSIONS:This is the first study to identify genes downregulated in LS-US compared to non-LS-US. Three genes were found to be both targets of LS-US associated miRNA and downregulated in LS tissue. Studies are being performed to corroborate the direct interaction of the miRNA and gene targets, along with cell-based assays to identify the potential molecular mechanism or role of these genes in LS. Overall, TGFB2 seems to be a key player in the development of LS. This information could allow us to better understand the pathophysiology of LS to better direct non-invasive therapies.