A Comprehensive Pan-Cancer Gene Expression And Drug Sensitivity Analysis Reveals Slfn11 As A Marker Of Sensitivity To Dna-Damaging Chemotherapy
Kevin Shee, PhD1, Lael S. Reinstatler, MD, MPH2, John D. Seigne, MD2, Todd W. Miller, PhD1
1Geisel School of Medicine at Dartmouth, Lebanon, NH, 2Dartmouth Hitchcock Medical Center, Lebanon, NH
BACKGROUND: Precision medicine seeks to integrate data from a patientís cancer to effectively tailor anti-cancer therapy. DNA-damaging chemotherapies have been successfully used to treat urologic cancers, but tend to be associated with significant toxicities. Thus, the identification of biomarkers of response to chemotherapeutics may allow providers to limit treatment to only those patients who stand to clinically benefit. Here we have developed and implemented a comprehensive pan-cancer analysis integrating gene expression and drug sensitivity profiles to identify novel biomarkers of response to DNA-damaging chemotherapeutics.
METHODS: Gene expression profiling and drug sensitivity analyses from the Cancer Cell Line Encyclopedia (CCLE) database (860 cell lines x 481 drugs), the Genomics of Drug Sensitivity in Cancer (GDSC) database (1065 cell lines x 266 drugs), and the National Cancer Institute 60 database (60 cell lines x 237 drugs) were correlated using Pearsonís method. 1-way ANOVA was used to compare relationship between chemotherapeutic classes. Human tumor gene expression and protein data was obtained from the Human Protein Atlas.
RESULTS: The only gene found to be significantly correlated with drug sensitivity in correlation analysis for all 4 classes of chemotherapeutic agents in all 3 databases was SLFN11, or Schlafen Family Member 11. Cancer cells with high SLFN11 gene expression were found to be highly correlated with sensitivity to DNA-damaging chemotherapeutic agents (Fig 1A; p<0.0001). This relationship was not found for similar analyses performed for microtubule inhibitors (p>0.05). When stratifying the analysis by chemotherapeutic class, SLFN11 expression was found to be an especially strong marker of topoisomerase inhibitor sensitivity (Fig 1B; p<0.05 by 1-way ANOVA for each dataset). RNA sequencing and IHC for SLFN11 gene expression and protein, respectively, in human tumors showed highest levels of expression in renal cell tumors among all tumor types.
CONCLUSIONS: Using pan-cancer cell line gene expression profiling and drug sensitivity data, we have identified SLFN11 expression as a novel biomarker of sensitivity to DNA-damaging chemotherapeutics, particularly topoisomerase inhibitors, that is highly expressed in renal cell tumors. 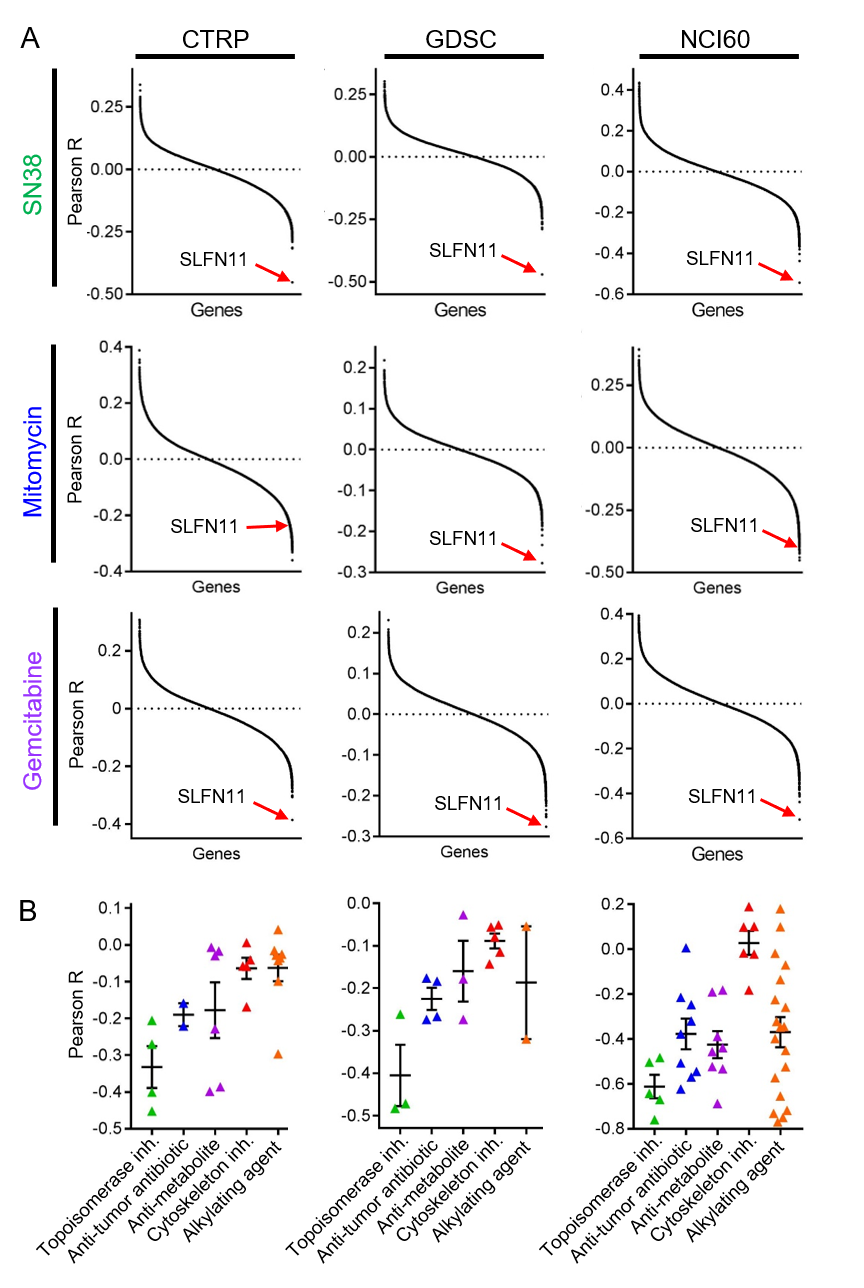
Back to 2019 Abstracts
