Identifying Gene Expression to Predict Biochemical Recurrence Following Radical Prostatectomy
John Corradi, PhD, Christine Cumarasamy, MD, Ilene Staff, PhD, Joseph Tortora, MS, Andrew Salner, MD, Joseph Wagner, MD
Hartford Hospital, Hartford, CT
BACKGROUND
Identification of novel biomarkers associated with high risk prostate cancer or biochemical recurrence can drive improvement in detection, prognosis, and treatment. The current understanding of prostate cancer genomes is still emerging, and studies can be limited by small sample sizes and sparse clinical follow-up data. We utilized a large sample of prostate specimens to identify gene expression to predict biochemical recurrence following radical prostatectomy.
METHODS
Between 2008 and 2011, patients undergoing radical prostatectomy at Hartford Hospital were consented to submit specimens for whole genome gene expression as part of the Total Cancer Care Consortium. RNA isolated from formalin-fixed paraffin-embedded prostates was hybridized to a custom Affymetrix microarray. Regularized (LASSO) Cox regression was performed with cross-validation to identify a gene expression signature that improves risk prediction. Recurrence was defined as post-operative PSA >0.2 ng/mL or triggered salvage treatment. Model performance was assessed using time-dependent ROC curves (with area under the curve, AUC) and survival plots.
RESULTS
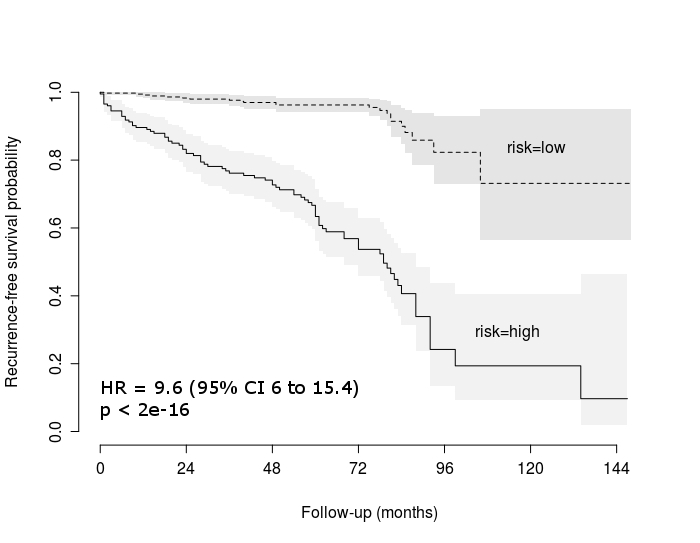
Pre- and post-operative PSA data were available for 606 prostate specimens. Using the LASSO model, a 5 gene signature was identified that independently predicted biochemical recurrence above Gleason grade and tumor stage. The time-dependent ROC AUC for the 5 gene signature with Gleason grade and tumor stage was 0.868 compared to an AUC of 0.767 for Gleason grade and tumor stage alone (first figure below). Patients stratified into low and high risk groups based on the predictive model score displayed significant differences in their recurrence-free survival curves (second figure below). The predictive model was subsequently validated on two independent gene expression data sets. The model included genes (RHOU, MTX2, and ERP44) that have previously been implicated in prostate cancer biology.
CONCLUSIONS
Our unique 5 gene signature panel can improve prediction of biochemical recurrence over the use of classical pathological hallmarks alone. Further research should validate the 5 gene signature in more specific sub-populations of prostate cancer patients, including those with earlier biochemical recurrence. 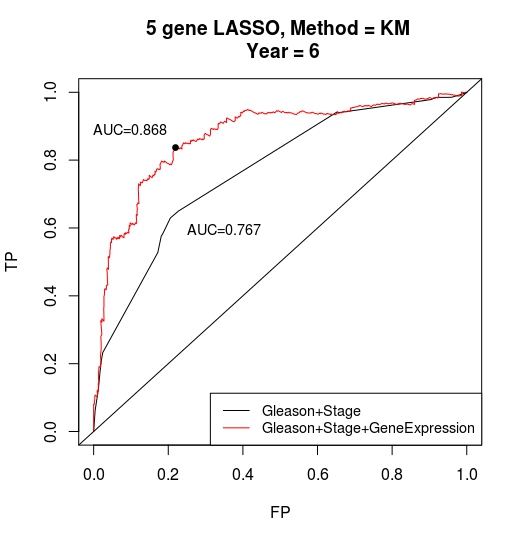
Back to 2019 Abstracts
