Metabolomic Evaluation of MRI-US Fusion Biopsies Differentiates Malignant from Benign
Adam S. Feldman, MD, MPH; Lindsey Vandergrift, BA; Taylor Fuss, BA; Emily Decelle, BA; Shulin Wu, PhD, Chris Dietz, PhD, Felix Ehret, PhD, Sarah Dinges, BA; Yannick Berker, PhD, Chin-Lee Wu, MD; PhD, Leo Cheng, PhD
Massachusetts General Hospital, Boston, MA
BACKGROUND: Multiparametric MRI (mpMRI) has improved detection of prostate cancer (PCa), however, inaccuracies remain. Our prior work with ex vivo magnetic resonance spectroscopy (MRS) demonstrated metabolomic field differences in PCa and benign prostate tissue. This study uses ex vivo MRS to study mpMRI/ultrasound fusion targeted biopsies to identify metabolomic markers of disease.
METHODS: Subjects having fusion biopsy were eligible for the study. ≥3 targeted cores were taken, followed by standard 12 core template biopsy. Patient matched target and nontarget cores (n=54 patients) underwent high resolution magic angle spinning MRS on a Bruker 600MHz spectrometer. Spectra were processed and transformed into statistical matrices using a MatLab program. Principal component (PC) analysis was performed with all identified metabolic spectral regions (n=33). Following MRS, all biopsy cores underwent standard histopathology.
RESULTS: 59% of fusion targets were positive for PCa. Higher PIRADS scores resulted in greater likelihood of detecting PCa. Patient matched target and nontarget samples were compared with Wilcoxon tests. Among the 33 metabolic spectral regions, ten regions differentiated between all paired samples with statistical significance. The Figure demonstrates a heatmap of metabolic regions measured in targeted cores. Although there were minor differences between cores from PIRADS 4 & 5 lesions, the majority of metabolic regions were similar. The second data column in the heatmap demonstrates significant differences between the analyzed (MRS) cores that were PCa-positive vs. cores that were benign. The third data column shows that similar differences remain present even when the analyzed target core is benign, but other cores from that MRI target are PCa-positive. These findings support the presence of metabolomic fields in PCa tissue.
CONCLUSIONS: We demonstrate metabolic differences in tissue obtained from mpMRI lesions as compared with mpMRI normal regions. Metabolic differences are also seen in mpMRI lesions with biopsies demonstrating cancer as compared with those lesions from which all biopsies were benign. Continued investigation of metabolic differences in mpMRI lesions may help translate ex vivo MRS to novel in vivo MRS biomarkers and thus improve the discrimination of PCa from normal with imaging. 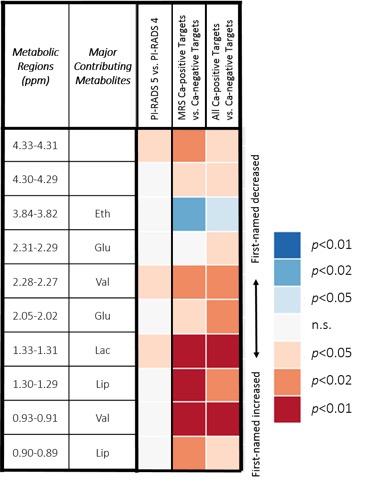
Back to 2018 Program
