| NEAUA Main Site | | | Past & Future Meetings |
|

|
Genomic Heterogeneity and the Small Renal Mass Daiki Ueno, MD1, Marta Boeke, PhD1, Jamil S. Syed, MD1, Kevin A. Nguyen, MS1, Patrick Macgillivray, MD2, Adebowale Adeniran, MD3, Peter Humphrey, MD PhD3, Yuval Kluger, PhD3, Zhonzhi Liu, PhD3, Harriet Kluger, MD4, Brian Shuch, MD1. 1Yale Department of Urology, New Haven, CT, USA, 2Yale Department of Molecular Biophysics and Biochemistry, New Haven, CT, USA, 3Yale Department of Pathology, New Haven, CT, USA, 4Yale Department of Medicine, New Haven, CT, USA.
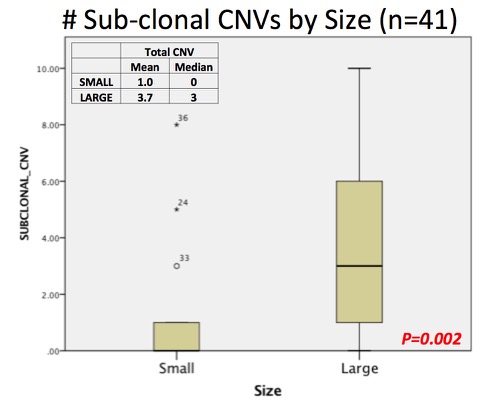
BACKGROUND: Pre-treatment genomic characterization of the small renal mass is now feasible, however extensive tumor heterogeneity in renal cell carcinoma (RCC) may represent a barrier to widespread adoption. This concept emerged from multi-site assessment of large renal masses. We set out to evaluate genomic heterogeneity in resected small and large renal tumors to provide further insight into the limitations of this approach. METHODS: A consecutive series (n=100) of nephrectomy specimens had 3+ regions sampled >1 cm apart at the time of gross pathologic exam. A total of 47 small (cT1a) and large (cT2+) clear cell tumors were selected for evaluation. DNA was extracted for copy number variation (CNV) of common driver alterations from the Cancer Genomic Atlas (TCGA) using an Illumina Human CytoSNP12 array. Gene expression analysis was performed with a custom Nanostring digital RT-PCR array and analyzed with nSolverAnalysis Software to characterize ccA vs ccB profiles as well as the Prolaris Cell Cycle Progression (CCP) score. Total and subclonal CNVs, CCP score, and ccA/B classification was assessed by tumor, size grouping, and individual region. Back to 2017 Program |
|
Photos courtesty of Old Port of Montréal © Tourisme Montréal, Stéphan Poulin.